Cardiac Electrophysiology Web Lab
This is 'Web Lab 2', a second generation Web Lab incorporating experimental data and parameter fitting. You can read more about our plans in a preprint of our Web Lab 2 paper.
Quick start links
- View results of experiments stored on this site.
- Compare the results of different experiments, e.g. dog models under 2Hz pacing, an IV curve of the fast sodium current, or S1-S2 and steady state restitution; or set up your own comparisons.
- View prototype parameter fitting results.
- Analyse your own models/protocols.
- Find out more about this site - read on!
What is the Web Lab?
Modellers have adopted XML-based markup languages to describe mathematical models over the past decade. This is great, as it means that models can be defined unambiguously, and shared easily, in a machine-readable format.
We have been trying to do the same thing with 'protocols' - to define what you have to do to replicate/simulate an experiment, and analyse the results. We can then curate models according to their functional behaviour under a range of experimental scenarios.
For the first time, we can thus easily compare how different models react to the same protocol, or compare how a model behaves under different protocols.
This website is our prototype Web Lab for the domain of cardiac electrophysiology. It brings together models encoded using CellML and virtual experiment protocols encoded in our own language, using standardised tags to generate interfaces between them, doing all the necessary units conversions. The stored results of these experiments can be viewed and compared.
If you wish to analyse your own models or create new protocols, you will need to register for an account and have it approved by a site admin.
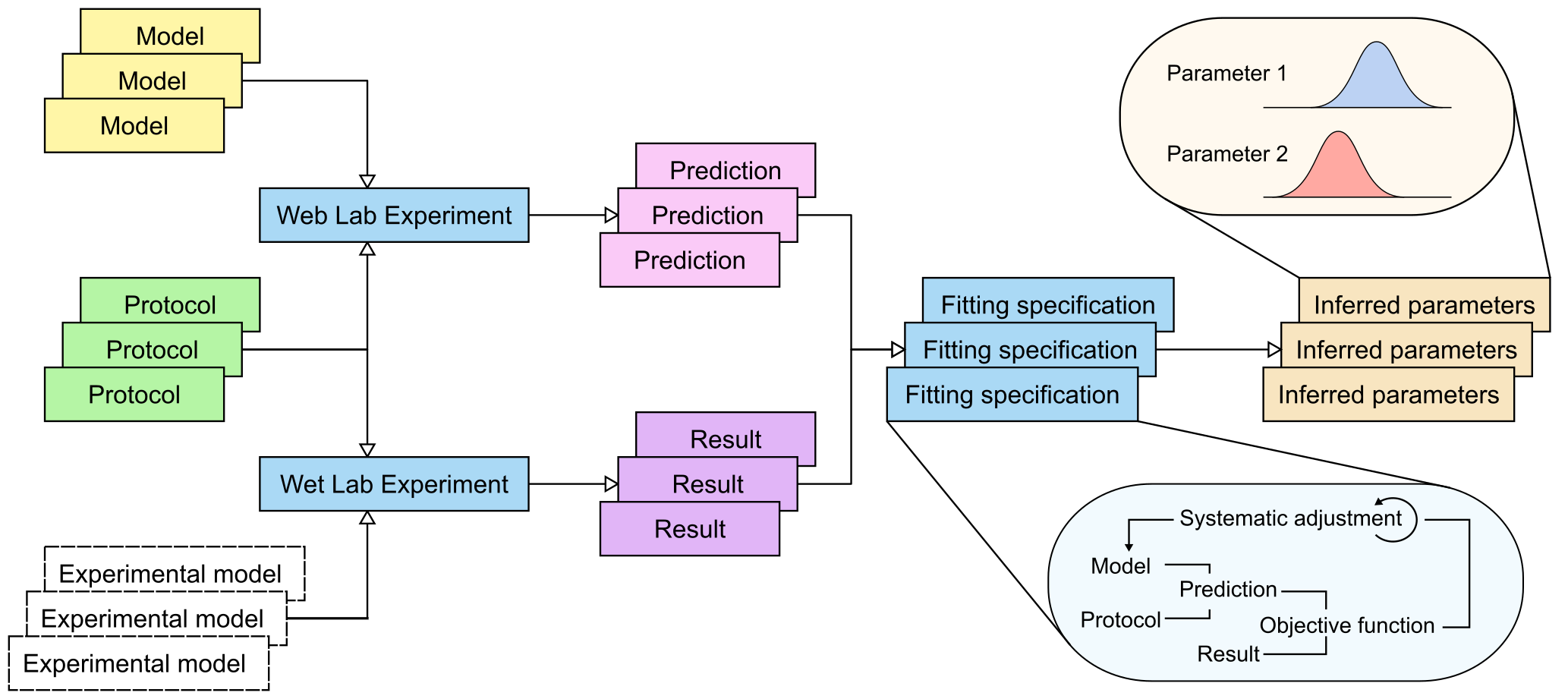
This website will store models, protocols and results (from both wet lab and in silico experiments), allowing comparisons to be made, and model parameters to be fit to data.
Have a look at the main experiment database to get started, or read more about the system.